Protein liquid-liquid phase separation (LLPS) can drive the assembly of various membrane-less compartments involved in a wide range of biological processes. An increasing number of proteins with diverse biological functions have been reported to undergo LLPS both in vitro and in vivo. Protein LLPS is highly sensitive to the conditions such as protein concentration, pH, ionic strength, metal ions, temperature, RNA, and crowding agent. Many fundamental questions remain to be answered. For instance, how to effectively find the LLPS condition of a given protein? How to compare the LLPS ability of different proteins? Moreover, protein encounters a highly complicated cellular environment in vivo. The LLPS behavior of each individual protein is most likely distinct from that in cells. Therefore, it is important to investigate whether and how protein LLPS is dynamically regulated in the presence of native binding partners, cofactors, PTMs, and mutations. On February 12th, a research article titled “A high-throughput method for exploring the parameter space of protein liquid-liquid phase separation” was published on the Cell Reports Physical Science journal by Prof. LIU Cong group from the Shanghai Institute of Organic Chemistry, Chinese Academy of Sciences, and Prof. LI Dan group from Shanghai Jiao Tong University. In this paper, the researchers established a high-throughput protein phase separation (HiPPS) profiling method to evaluate the LLPS ability of different proteins. Screening results reveal that protein LLPS is a general property of both well-folded and unstructured proteins. More importantly, the ability of protein LLPS (termed as LLPS space) is under the regulation of protein-protein interaction, posttranslational modification, genetic mutation and chaperones. In this study, the researchers established HiPPS contained 96 conditions based on the factors that may influence protein LLPS. During the screening process, a crystallization robot and a high-content analysis systems were used to quickly and efficiently monitor protein LLPS while minimizing the use of protein samples. Screening results suggested that LLPS ability of proteins is intimately contingent on conditions, and LLPS appears to be a general property of both unstructured and well-folded proteins. Notably, multi-component co-LLPS mixture decreases the threshold concentration of individual proteins for LLPS, and LLPS space of protein can be dynamically reshaped by protein-protein interaction, posttranslational modification, genetic mutation, and cofactors (Fig.1). Together, this work suggests HiPPS method may effectively characterize LLPS of individual proteins or protein mixtures in vitro, which is valuable for understanding its role in biological processes (Fig. 1). 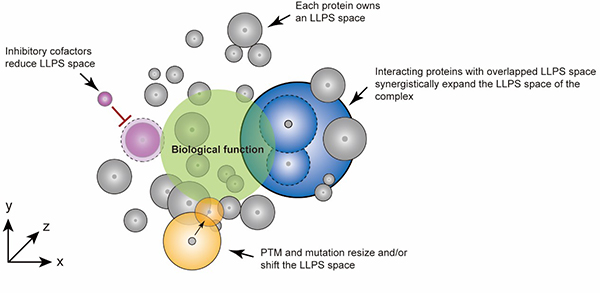 Fig.1 Characterization and dynamic regulation of protein LLPS space(Image by LIU Cong) LIU Cong Ph.D. Professor Tel: 021-68582528 Email: liulab@sioc.ac.cn |


